Novel Target Discovery via Reverse Immunology
Dec 20, 2022
Target-based drug discovery – looking for Holy Grails of pharmacology
Target-based drug discovery is the most common strategy for developing new medications. However, when it comes to clinical trials, most new drugs fail due to inadequate cure rate (efficacy) or safety concerns. The extremely low success rate – only about 12% of drugs make it to the market – suggests that a “wrong” target has been selected during the target discovery process.
Identifying the molecules and mechanisms primarily involved in disease is an essential first step in discovering targets that can be acted upon therapeutically. This initial step traditionally begins with a manual investigation of scientific literature and biomedical databases to gather evidence linking particular proteins to disease. This quest relies on using bioinformatics tools and collecting experimental evidence supporting the hypothesis that changing the molecule’s function will affect a patient’s health.
Drug target – what is it, exactly?
A drug target can be defined as a molecule in the body, usually a protein, that is intrinsically associated with a particular disease. A perfect drug target should be involved in a crucial biological pathway, be functionally and structurally well characterized, and be druggable – capable of binding to therapeutic molecules.
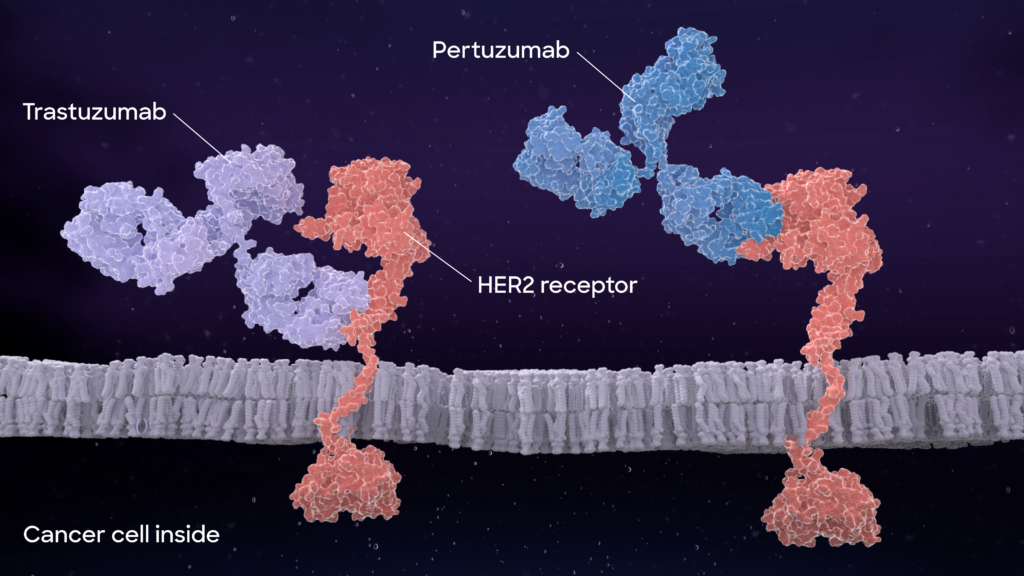
In a perfect scenario, a drug (here a therapeutic antibody) should bind only to the intended target and initiate its structural or functional change for therapeutic effect.
It sounds simple, but in reality, it is one of the most challenging ventures undertaken by humanity. The search for new pharmacological targets is often hampered by partial information about disease pathogenesis, usually consisting of conflicting data. Much knowledge about molecules in our bodies is based on indirect evidence from animal, cell, and tissue culture experiments. Unfortunately, therapies based on animal model biology often fail to replicate when tested in human trials. Moreover, many proteins are druggable based on their structure – they will bind the drug – but this will not lead to therapeutic benefit.
Researchers also rely on human epidemiological studies, for example, comparing blood levels of assumed disease-related molecules. Still, these results are subject to erroneously assigning cause and effect or misidentifying correlation for causation. Thus, there’s a desperate need for more robust information about targets and their potential clinical efficacy.
Why can’t we make more drugs?
The current approach of target-based drug design is not limited by the technical hurdles to produce the drugs but by the need for progress in identifying novel biological targets. In theory, all biological macromolecules – about an estimated 10,000 – are potentially druggable targets, but only about 400 are currently approved. Over the past 50 years, the number of drugs sharing the exact mechanism of action or target has averaged about 4, ranging from 2-14 drugs per single target. So, it seems companies struggling to identify new targets fight over those recognized as therapeutically beneficial, trying to make a better mouse trap than their competitors.
In recent years, human genomics has been increasingly revealing its power. The underlying idea is to use the powerful tool of human genetic association to identify key drug targets involved in the disease, which can be changed or blocked by drugs. However, these approaches require sifting through data from mega-repositories and large case-control consortium-type studies. The biggest challenge here is the data originating from multiple sources with unclear predictability and consistency. In addition, those associations require further studies to resolve casual relationships to confirm that the putative targets are indeed involved in disease pathogenesis and susceptible to therapeutic modulation.
Can we learn from our bodies how to pick drug targets?
Our bodies can fight and cure diseases, including cancer and infections. Could we investigate how our immune systems pick the targets to apply self-therapy? Absci is pioneering the discovery of new drug targets utilizing a process we call reverse immunology. Our approach is grounded in natural immunology, and we use our proprietary, integrated wet lab screening and deep learning to open the door to new drug target identification.
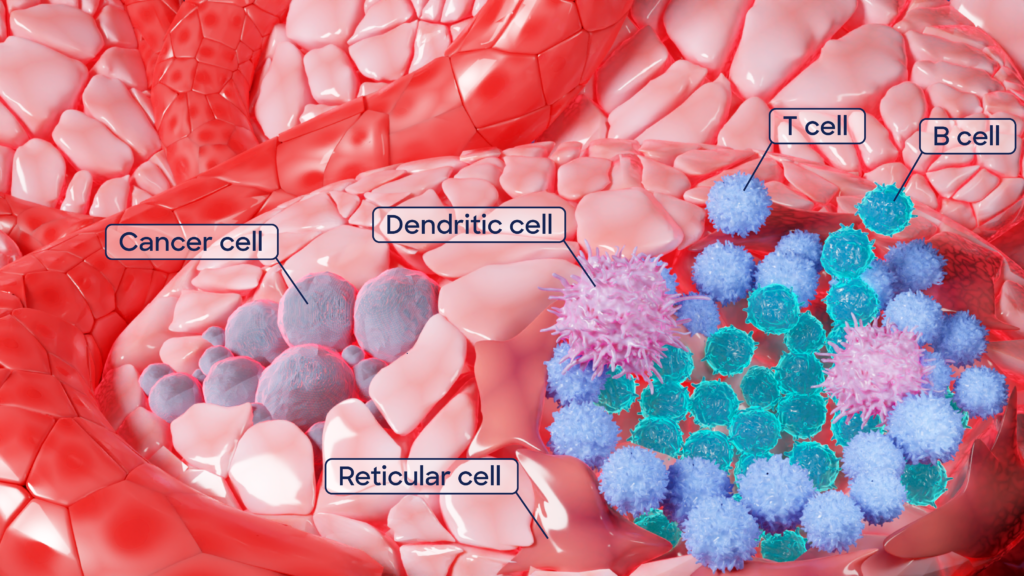
How does reverse immunology work? It starts with patient samples; these could be a patient population that displays exceptional immune responses in studies. The secret to the new targets lies in a particular tissue, called tertiary lymphoid structures, or TLSs. They resemble lymph nodes – key hubs of immune cell manufacturing – however, they don’t form in healthy tissue but only in places with chronic inflammation, infection, transplanted organs, autoimmunity, or cancer. How they develop is still under investigation, but they are most likely our body’s response to long-lasting exposure to inflammation caused by unhealthy conditions. TLSs are embedded special forces of the immune system, deployed to disease-affected tissues to produce defenses locally. A small army that identifies the enemy and delivers the ultimate weapon, target-specific antibodies, against the disease-causing cells or pathogen.
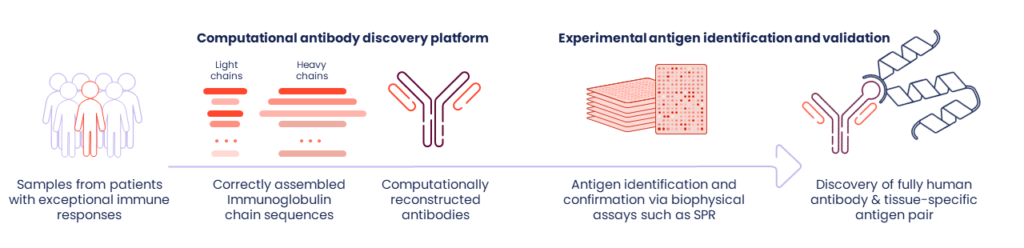
We call our approach “reverse immunology” because we start with those immune cells from TLSs and identify the antibodies they produce in the greatest quantities. What we are looking for, specifically, are the antibody blueprints from immune cells – RNA sequences, which we assemble computationally. We then use those antibody sequences to identify target antigens – rather than starting with a target and generating or fishing for an antibody. Our process is then reversed.
After reconstructing fully human antibody sequences in silico, we produce these antibodies in the lab. We use high throughput proteomic screening methods to determine their target antigens in an experimental process we call “deorphaning.” At the end of this reverse immunology process, we have the immunologic target paired with a fully human monoclonal antibody that can be used as a starting point for drug development.
Our methods are exceptionally robust – they do not require the isolation of single immune cells or the processing of fresh tumor tissues. Instead, we can work with RNA-seq data we generate from banked tissue samples, including archival specimens collected by academic consortia, clinical trials, and commercial biobanks. Thus, we can direct our technology toward curated source tissues selected for desired disease and therapeutic response profiles.
We have constructed an extensive library of antibody-target pairs. Some antigens we’ve found are the usual suspects already known to be either essential mediators of immuno-oncology responsiveness in the case of cancers or neutralizing antibodies in the case of SARS-CoV2. Importantly, we’ve also identified many novel antigens that may have promise as therapeutic targets, and we have fully human antibodies in hand.
Absci’s reverse immunology approach is powerful and efficient, offering an unbiased antibody and target discovery method. We believe it will allow us to create better biologics for patients faster.